Patient-specific whole brain blood flow simulations
We are concerned with performing neurovasculature blood flow simulations in support of clinical neurosurgery. Cerebral blood flow behaviour plays a crucial role in the understanding, diagnosis and treatment of cardiovascular disease; problems are often due to anomalous blood flow behaviour in the neighbourhood of bifurcations and aneurysms within the brain. Simulation offers the possibility of performing patient-specific, virtual experiments to study the effects of courses of treatment with no danger. For this work, we will use our Lattice-Boltzmann code, HemeLB[4,3], designed to simulate fluid flow in the sparse topologies of the human brain. The code can create visualizations from within a running simulation and send them to a viewing client on a workstation situated in, eventually, the hospital. The clinician can then steer the parameters of the simulation.
We will also utilize an advanced international grid infrastructure that we have put in place to allow clinicians to seamlessly interact with some of the worlds biggest supercomputers from their operating theatre, and to access patient-specific haemodynamics simulations in real-time.
Overview
Cerebral blood flow behaviour plays a crucial role in the understanding, diagnosis and treatment of neurovascular pathologies, where problems are often due to anomalous blood flow behaviour in the neighbourhood of bifurcations and aneurysms within the brain, although the details of such pathologies are not well understood. The aim of this project is to use fluid flow simulation in combination with patient specific medical imaging data in order to not only help understand blood flow patterns in the presence of neurovascular pathologies, but also to provide a tool which surgeons can use in diagnostic and therapeutic capacities.
Using blood flow simulation to diagnose and predict the outcomes of treatment of intracranial vascular pathologies is an area of patient specific medical simulation that requires the power of thousands of cores, possibly spread over multiple computational resources. This is particularly true where the haemodynamics through the entire intravascular arterial tree needs to be simulated within clinically relevant time frames. To treat aneurysms and arterial-venous malformations (AVM), image guided endovascular surgery, which is also known as interventional neuroradiology, can be used. This intervention through the application of, for example, an embolisation to rectify the blood flow can substantially alter the haemodynamics in terms of flow rates and pressures throughout the vasculature. Having a computational tool available which surgeons can use, not only to examine the pressure and velocity variations through the vasculature, but also to predict what changes might occur as a result of surgical intervention, would be an invaluable addition to their surgical tool kit.
Computational Approach
This work uses the HemeLB code - which is well-suited to describe fluid behaviour - within sparse systems such as neurovasculatures. The code is written in C/C++ and parallelized using MPI. It employs a number of algorithmic optimizations and techniques to efficiently compute and communicate flow data in sparse and complex geometries. The domain decomposition ensures that computational domains are equally partitioned. This, combined with overlap of communication with interior fluid site computation results in super-linear speedup over hundreds of cores. Efficient layout of memory and memory access patterns further optimizes the fluid simulation.
HemeLB is also designed to efficiently run in cross-site scenarios such as with MPIg, where the domain is decomposed such that intermachine communication is minimized and intermachine communication is overlapped with intramachine communication. HemeLB has built in real-time rendering and steering capabilities such that the time-varying behaviour of the blood flow can be visualized live by a parallel in situ ray tracing algorithm. Each frame is transmitted over the network to a workstation which visualizes the frame in real time. The visual and physical parameters can be interactively modified through steering capabilities. The analysis and visualization, particularly in real-time, is essential to this successful use of this code in clinical environments.
Acknowledgements
This work is funded by UK EPSRC (Grant No EP/F00561X/1) within the joint US-UK project called GENIUS (grid-enabled neurosurgical imaging using simulation) and supported by RealityGrid Platform Grant (Grant No EP/C536452/1), EU Virtual Physiological Human project (Reference FP7-ICT-2007-5.3) and Virolab (EC 027446).
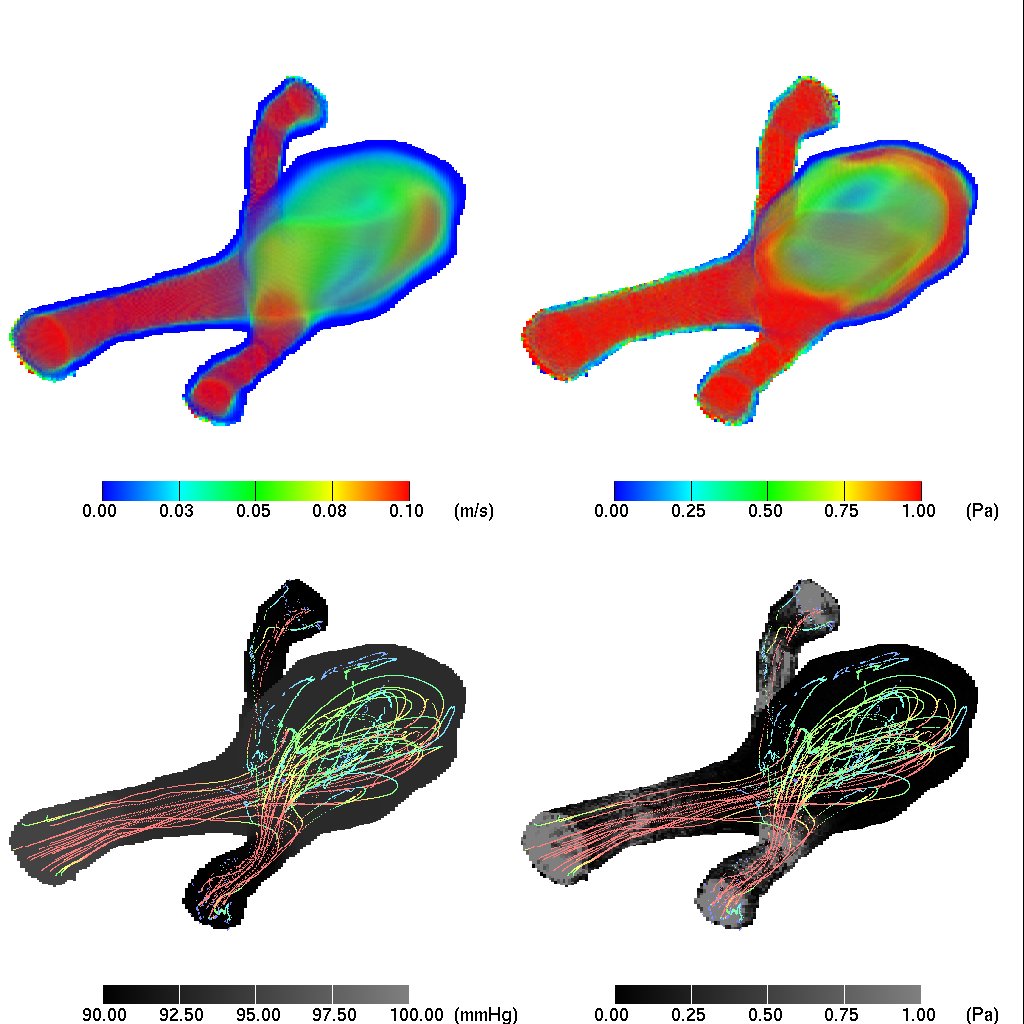
Fig.1: A visualization of an aneurysm. (Top left: volume rendered velocity; top right: volume-rendered von Mises stress; bottom left: external pressure and streaklines; bottom right: external von Mises stress and streaklines).
References/Literature
[1] Doctors et al. A computationally efficient method for simulating fluid flow in elastic pipes in three dimensions. Computer Physics Communications (2010) vol. 181 (9) pp. 1584 - 1592. 10.1016/j.cpc.2010.05.012
[2] Manos et al. Patient specific whole cerebral blood flow simulation: A future role in surgical treatment for neurovascular pathologies. Proc. of Teragrid (2008)
[3] Mazzeo et al. In situ ray tracing and computational steering for interactive blood flow simulation . Computer Physics Communications (2010) vol. 181 pp. 355-370 10.1016/j.cpc.2009.10.013
[4] Mazzeo and Coveney. HemeLB: A high performance parallel lattice-Boltzmann code for large scale fluid flow in complex geometries. Computer Physics Communications (2008) vol. 178 (12) pp. 894 - 914 10.1016/j.cpc.2008.02.013
[5] Sadiq et al. Patient-specific simulation as a basis for clinical decision-making. Phil. Trans. R. Soc. A (2008) vol. 366 (1878) pp. 3199-3219 10.1098/rsta.2008.0100